MSA220/MVE440 Exam
Exploratory data analysis
探索性数据分析代写 The exploratory data analysis shows centred values around 0 with relatively controled standard deviation closed to 1. Thus…
mean per feature
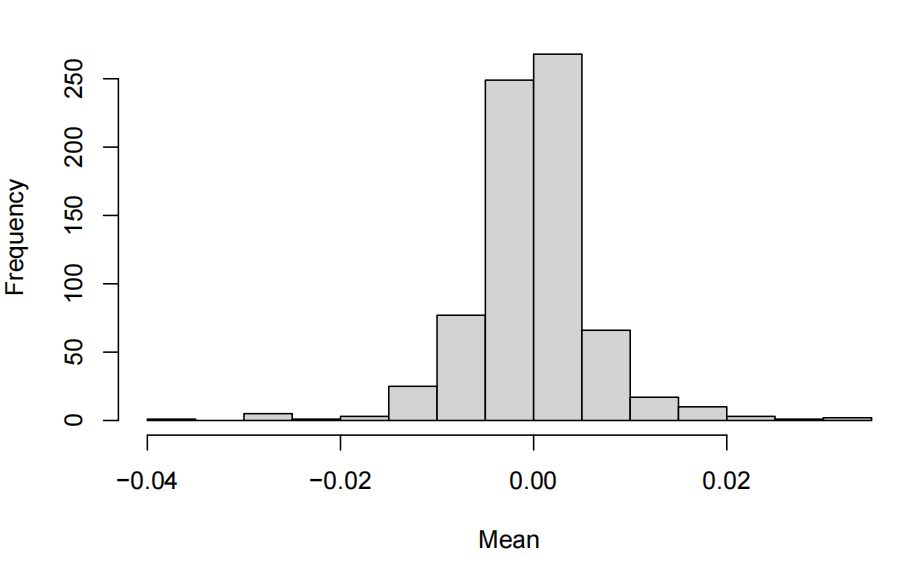
standard deviation per feature
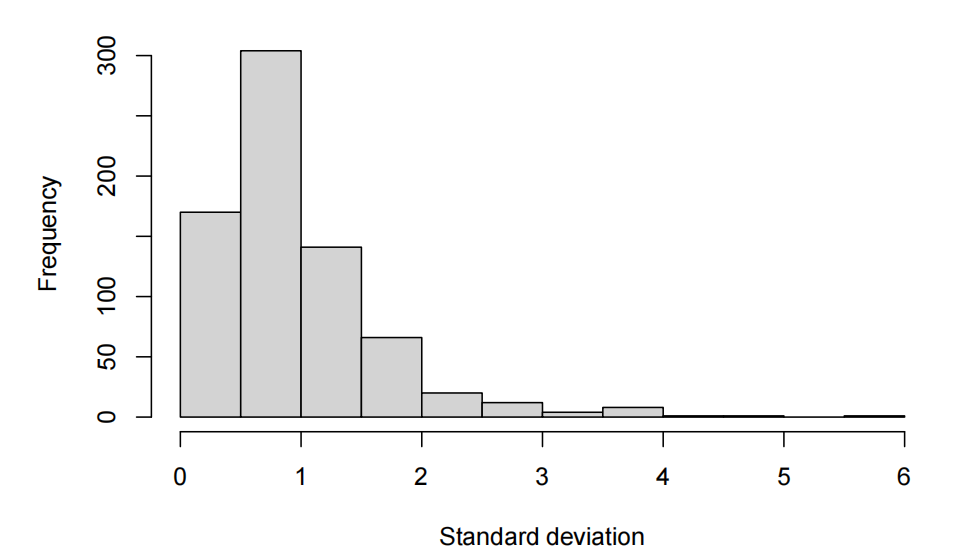
- The exploratory data analysis shows centred values around 0 with relatively controled standard deviation closed to 1. Thus, there is no need to standardize in case we want to use PCA.
Data visualization 探索性数据分析代写
- Plots of one feature vs another feature are not informative since we have 728 features to choose from. Dimension reduction such PCA or embeddings like t-SNE or UMAP are recommended. Note that some of these methods are just suited for VISUALIZATION ONLY.
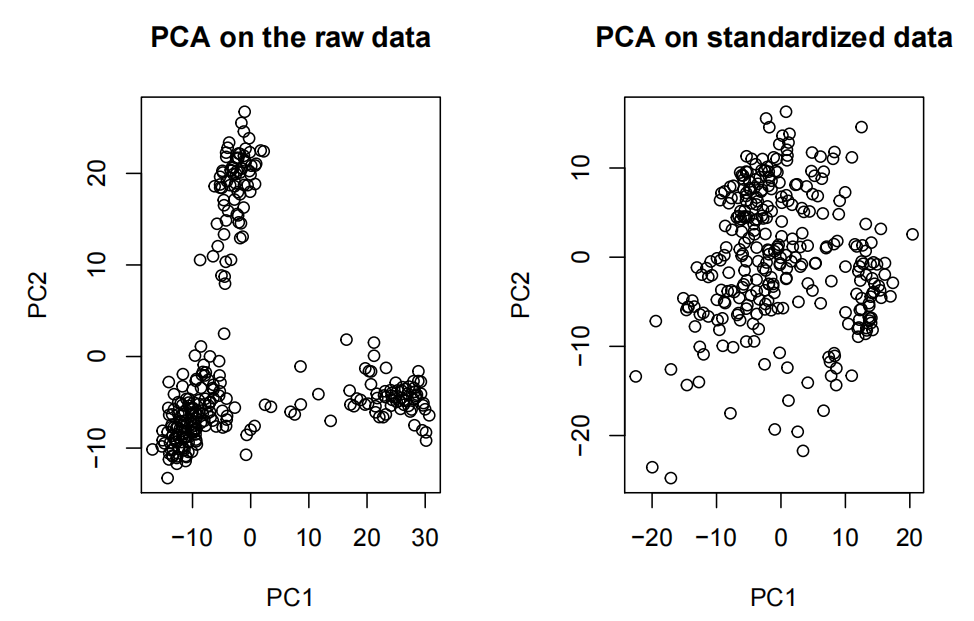
- Note the effffects of standardizing the data on the PCA.
- Note that saying, “I see 3 clusters on the PCA” is not enough to carry on with the analysis with predefifined k=3.
- Note that tSNE is defifinitely not recommended for clustering, just for visualzation.
Feature selection or dimension reduction before clustering 探索性数据分析代写
- Most clustering algorithms are not suited to handle high dimensional data. Thus, a method handling high dimensional data, dimension reduction (PCA) or feature selection (e.g. most variable features) before clustering is expected.
- Note again, tSNE is not recommended.
- Note, NMF is for non-negative matrices, if you use NMF you need to motivate how you handled the negative values!
- Important You need to defifine what is your imput matrix (and dimension) to the clustering algorithm, examples raw data with all the features, or the 10, 20 or 30 PC of the data (and motivation of why you chosed that many PC), most variable genes (how many and what was the criteria for selecting them).
Clustering 探索性数据分析代写
- Run an appropriate clustering algorithm, explain what parameters you used and motivate how you chose the parameters.
Selecting the number of clusters
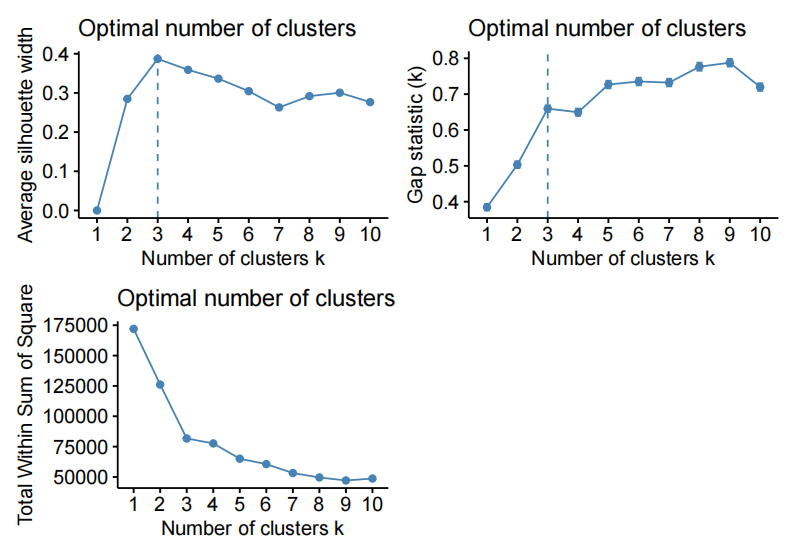
- Employ an analytical tool to select the optimal number of clusters, e.g. silhouette width, within sum of square error, between sum of square error, etc.
- The human eye on the dimension reduction is not the best tool.
- Note that “the elbow method” without saying what statistic the plot is showing is incomplete information.
- Note that there are statistics such as BIC that favor higher number of clusters, and thus, the “best” (higest or lowest) is not a correct answer, but “the highest drop” could be (elbow method on BIC)
## Loading required package: ggplot2 ## Welcome! Want to learn more? See two factoextra-related books at https://goo.gl/ve3WBa
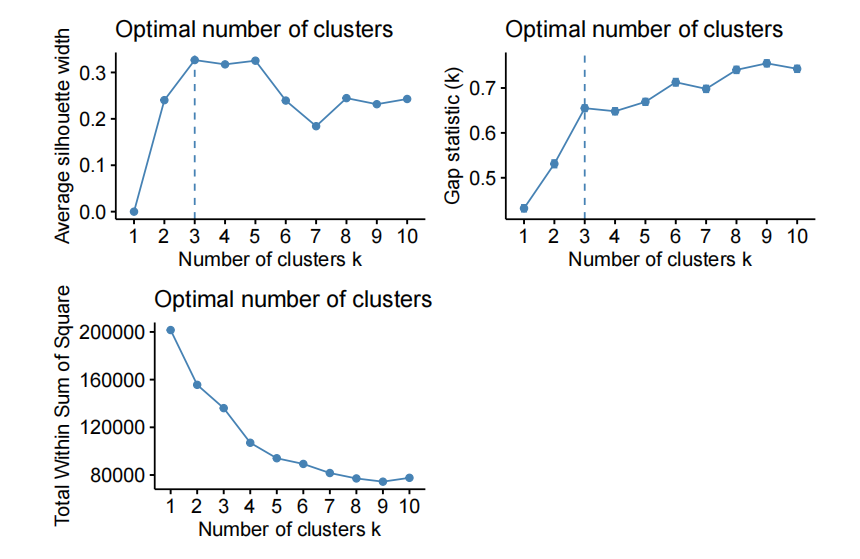
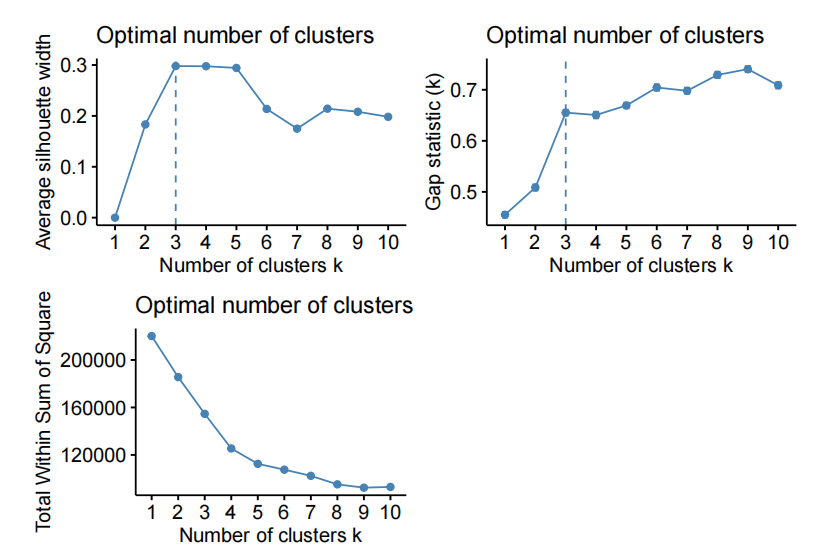
For example, varying the number of PC and using kmeans to cluster the data on the fifirst “n” PC of X.
- kmeans on the fifirst 10 PC of X
- kmeans on the fifirst 20 PC of X
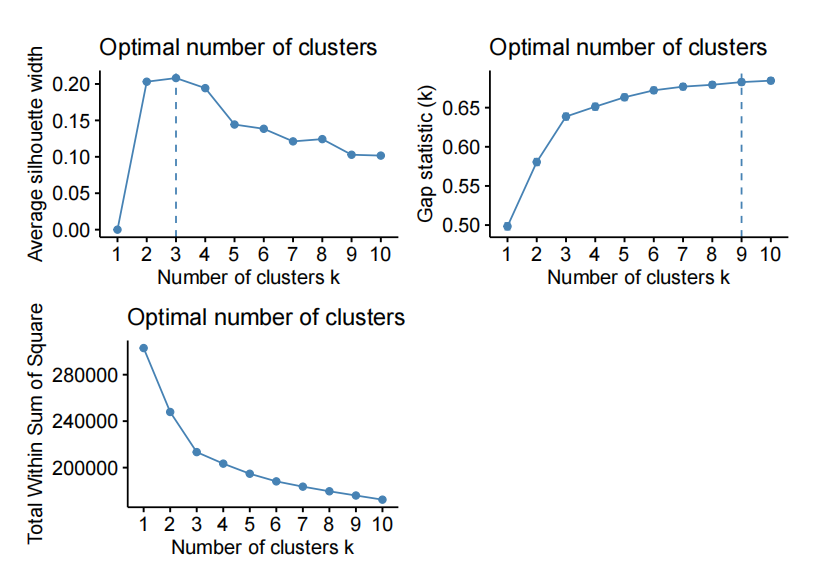
- kmeans on the fifirst 30 PC of X
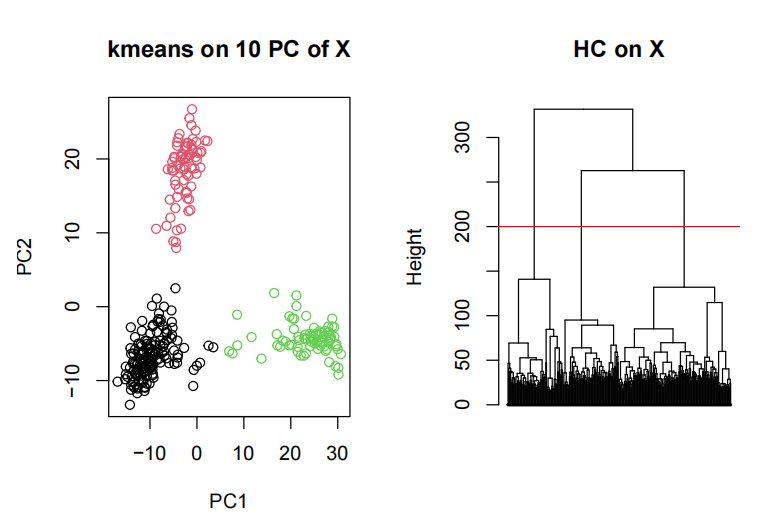
For exampel, HC on X with “ward.D2” distance
- Determine and motivate what is your optimal number of clusters
Visualize the results 探索性数据分析代写
You can use PCA or tSNE or any other embedding.
Optional
Measure the agreement between difffferent methods, e.g. ARI between kmeans and HC.
## Package ’mclust’ version 5.4.6
## Type ’citation("mclust")’ for citing this R package in publications.
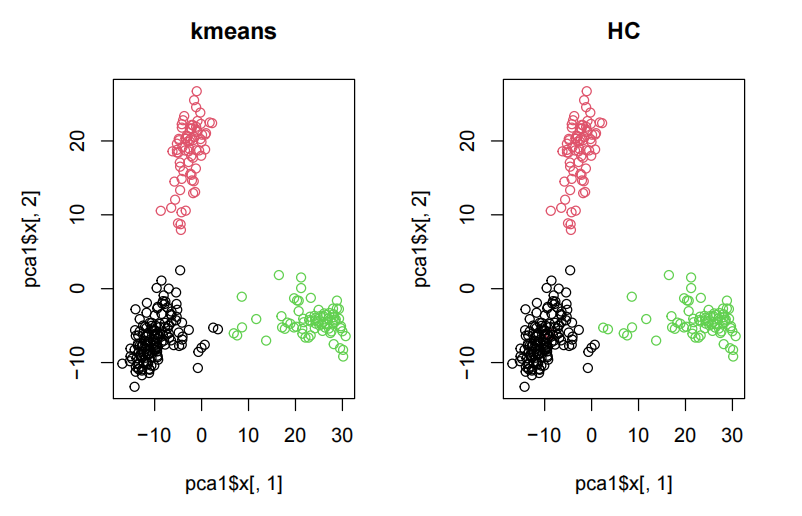
mclust::adjustedRandIndex(clusterCut, k3$cluster) ## [1] 0.9682521 Thus, kmeans and HC give almost the same results. par(mfrow=c(1,2)) plot(pca1$x[,1],pca1$x[,2], main = "kmeans", col=k3$cluster) plot(pca1$x[,1],pca1$x[,2], main = "HC", col=clusterCut)
par(mfrow=c(1,1))
Most important features per cluster
Once the clusters are assigned, you can use any statistical test to measure the difffference between two groups. Nevertheless, we are expecting you to use the methods learned in the course. For example, treating one cluster as class 1 and the rest of the cluster as class 0 (for each of the classes), taking the excercise as a classifification task and doing feature selection using e.g. sparse logistic regression, RF variable importance on bootstrap training data, etc. (see the comments on Excercise 1 for more information on feature selection), you could have exctracted the relevant features for each of the clusters.
Notice that you should motivate the method and the parameters you use and if you did CV, imbalanced data (if any), or any other thing relevant for that method.
Notice that we want the relevant features per cluster, not relevant features for the whole data. Thus, if you just mention the features relevant for the PCs, you need to justify how they are related to each cluster, although this type of answer is not prefered (see the the lines above).
Notice that for one cluster, the most variable features in that cluster is not a way to determine the relevant features for the cluster, we want variability between clusters, not within clusters.
Final notes 探索性数据分析代写
- Motivate motivate motivate, why you do the things you do (methods, dimension reduction, visualization, etc.), how you interpret the results, conclusions, etc.
- Be clear with what you do (methods, parameters, etc) and on what data you do it.
- Use captions, legends, titles, axis, anything that helps understanding the plots you are showing.



